Structural Complexity
Sebastian Ahnert
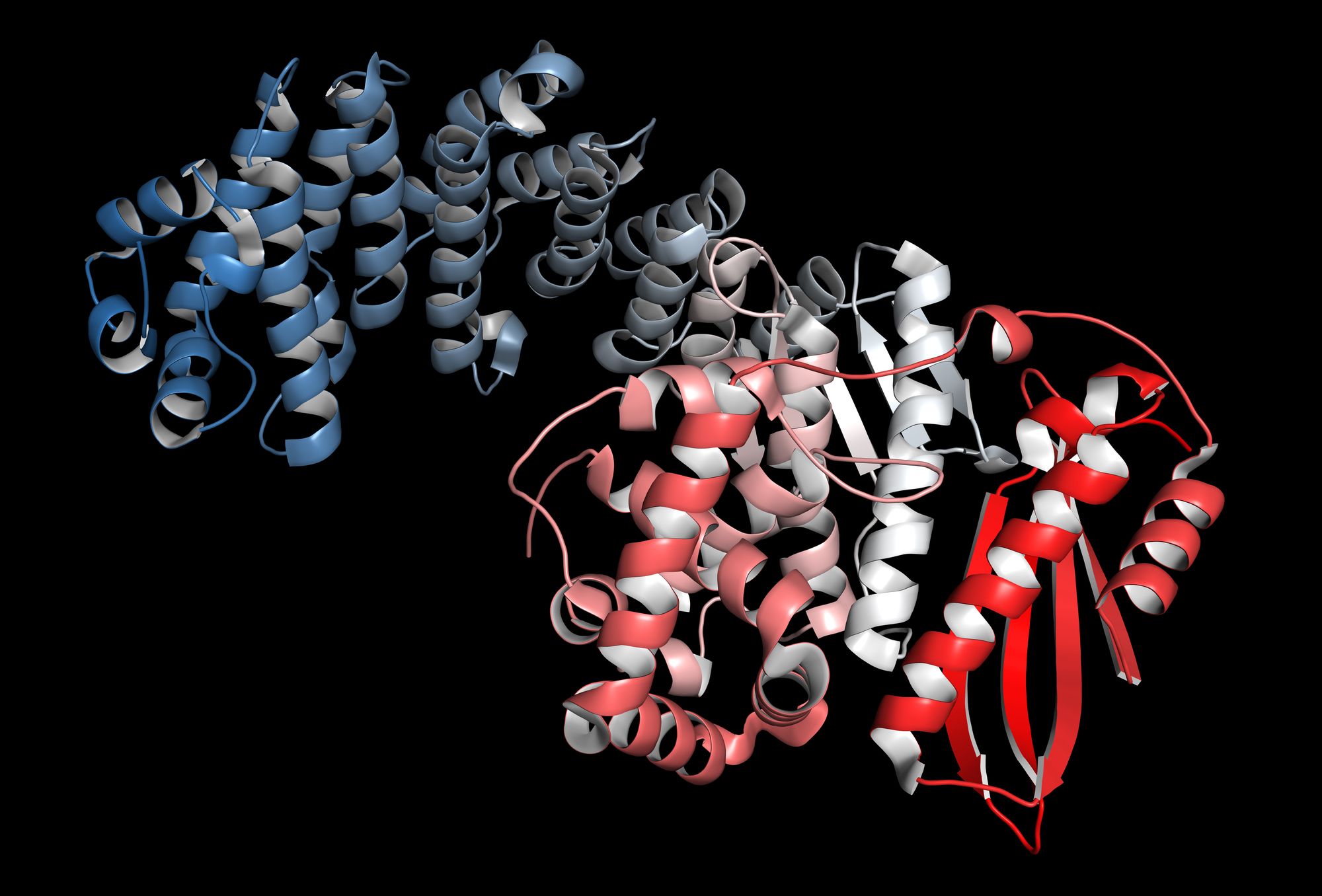
We are interested in studying, modelling, quantifying, and classifying structural complexity in a wide range of contexts, from biomolecules to self-assembling systems and networks. To achieve this, we use computational and theoretical approaches such as algorithmic information theory and network analysis.
The fundamental research questions we are interested in include: How does structural complexity arise in biological evolution? How much information do we require to encode a self-assembling structure? What can the algorithmic compression of network structures tell us about the network?
Dr Sebastian Ahnert led an interdisciplinary research group as a Royal Society University Research Fellow (2009-2017) and Gatsby Fellow (2016-2020) at the Cavendish Laboratory and Sainsbury Laboratory at the University of Cambridge, before joining CEB in 2020.
Research areas
Protein secondary, tertiary, and quaternary structure
We are interested in classifying and quantifying the space of possible protein structures. Our work on the 'periodic table of protein complexes' (Science, 2015) uncovered fundamental principles that govern the formation of protein complexes (or 'quaternary structures'), which play a vast range of roles in every biological organism.
It also offered a way of classifying existing complexes and predicting the likelihood of ones that had not yet been observed. We are currently working on various projects that examine how quaternary structure, tertiary structure, and secondary structure of proteins depend on each other, how mutations affect the protein at each of these structural levels, how the space of possible protein structures can be mapped, and how biological evolution has filled this space of possible structures.
RNA structure and genotype-phenotype maps
Biological information is typically stored, transmitted, and processed in the form of sequences, such as DNA, RNA, and amino acid chains. The fundamental link between biological information and biological outcomes is the genotype-phenotype map, which relates the space of possible sequences to the space of possible outcomes. Biological evolution as a process of selection takes place upon this scaffold. One can think of evolutionary selection as the driver of change, and the genotype-phenotype map as the road network that determines the accessibility of different parts of sequence space. The genotype-phenotype map of RNA secondary structure can be studied at a large scale, which allows us to understand the effect of the genotype-phenotype map on evolutionary outcomes. We have shown that the genotype-phenotype map leads to counter-intuitive properties of fitness landscapes, and that so-called 'fitness valleys' in such landscapes are much rarer than previously thought (Nature Ecology & Evolution, 2022).
Self-assembly and algorithmic complexity
Algorithmic complexity theory asks, 'What is the shortest algorithm that produces a given set of data?'. This description of data or information in terms of an algorithm is a powerful concept, as highly complex structures can be built using a simple rule set, and biological complexity, in particular, is highly algorithmic in this sense. We have extended the idea of algorithmic complexity to structures and have explored how we can describe and compare structures using self-assembly rules. Building on this, we have also shown (PNAS, 2022) that the ubiquitous symmetry and modularity of biological structures arise as a result of the way biological sequences (genotypes) map to biological outcomes (phenotypes).
Compressibility and structural complexity of networks
The computational study of large, complex networks has been a rapidly expanding research area over the past two decades. We have made various contributions to this field, firstly by using a compression approach to map the large-scale organisation of complex networks, secondly by developing new ways of analysing directed networks, particularly in the context of gene regulatory networks, neural networks, and social networks, and thirdly by bringing the analysis of network structure to new disciplines, such as food science and the study of large historical correspondence networks (Cambridge University Press 2020, Oxford University Press 2023).
Our members
Group leader
Sebastian Ahnert
Postgraduate students
Paula Garcia Galindo (PhD)
Sung Soo Moon (PhD)
William Lowe (PhD)
Nicholas Katritsis (PhD)
External members and visitors
Yavor Novev (PDRA)
Runfeng Lin (PhD)
Former members
Nora Martin (PhD)
Marcel Weiß (PhD)
Alexander Leonard (PhD)
Will Grant (PhD)
Yuanyie Chen (PhD, co-supervised)
Alexander Johnston (MPhil)
Salvatore Tesoro (PhD)
Emma Towlson (PhD)
Sam Greenbury (PhD)
Pascal Grobecker (Part III Physics)
Elliot Vaughan (CET IIB project)
James Simkins (CET IIB project)
Fátima González (summer student)
Toby Baker (summer student)
Marijana Vujadinović (summer student)
Pranav Reddy (summer student)
Eniak Alarcón (summer student)
Giles Barton-Owen (summer student)
Laura Imperatori (summer student)
Robert Baldock (summer student
Publications
P. Garcia Galindo, S. E. Ahnert, N. S. Martin
The non-deterministic genotype-phenotype map of RNA secondary structure
Journal of the Royal Society Interface 20, 20230132 (2023)
V. Mohanty, S. F. Greenbury, T. Sarkany, S. Narayanan, K. Dingle, S. E. Ahnert, A. A. Louis
Maximum mutational robustness in genotype-phenotype maps follows a self-similar blancmange-like curve
Journal of the Royal Society Interface 20, 20230169 (2023)
E. Corcoran, M. Afshar, S. Curceac, A. Lashkari, M. M. Raza, S. Ahnert, A. Mead, R. Morris
Current data and modeling bottlenecks for predicting crop yields in the United Kingdom
Frontiers in Sustainable Food Systems 7, 1023169 (2023)
A. Karjus, M. C. Solà, T. Ohm, S. E. Ahnert, M. Schich
Compression ensembles quantify aesthetic complexity and the evolution of visual art
EPJ Data Science 12, 21 (2023)
E. Corcoran, L. Siles, S. Kurup, S. E. Ahnert
Automated extraction of pod phenotype data from micro-computed tomography
Frontiers in Plant Science 14:1120182 (2023)
K. Dingle, J. K. Novev, S. E. Ahnert, A. A. Louis
Predicting phenotype transition probabilities via conditional algorithmic probability approximations
Journal of the Royal Society Interface 19, 20220694 (2022)
S. F. Greenbury, A. A. Louis, S. E. Ahnert
The structure of genotype-phenotype maps makes fitness landscapes navigable
Nature Ecology & Evolution 6, 1742 (2022)
N. S. Martin, S. E. Ahnert
Thermodynamics and neutral sets in the RNA sequence-structure map
Europhysics Letters 139, 3 (2022)
N. S. Martin, S. E. Ahnert
Fast free-energy-based neutral set size estimates for the RNA genotype-phenotype map
Journal of the Royal Society Interface 19, 20220072 (2022)
I. G. Johnston, K. Dingle, S. F. Greenbury, C. Q. Camargo, J. P. K. Doye, S. E. Ahnert, A. A. Louis
Symmetry and simplicity spontaneously emerge from the algorithmic nature of evolution
Proceedings of the National Academy of Sciences 119, e2113883119 (2022)
P. Roszak, J. O. Heo, B. Blob, K. Toyokura, Y. Sugiyama, M. A. de Luis Balaguer, W. W. Y. Lau, F. Hamey, J. Cirrone, E. Madej, A. M. Bouatta, X. Wang, M. Guichard, R. Ursache, H. Tavares, K. Verstaen, J. Wendrich, C. W. Melnyk, Y. Oda, D. Shasha, S. E. Ahnert, Y. Saeys, B. De Rybel, R. Heidstra, B. Scheres, G. Grossmann, A. P. Mähönen, P. Denninger, B. Göttgens, R. Sozzani, K. D. Birnbaum, Y. Helariutta
Cell-by-cell dissection of phloem development links a maturation gradient to cell specialization.
Science 374, 6575 (2021)
N. S. Martin, S. E. Ahnert
Insertions and deletions in the RNA sequence-structure map
Journal of the Royal Society Interface 18, 20210380 (2021)
S. Manrubia, J. A. Cuesta, J. Aguirre, S. E. Ahnert, L. Altenberg, A. V. Cano, P. Catalán, R. Diaz-Uriarte, S. F. Elena, J. A. García-Martín, P. Hogeweg, B. S. Khatri, J. Krug, A. A. Louis, N. S. Martin, J. L. Payne, M. J. Tarnowski, M. Weiß
From genotypes to organisms: State-of-the-art and perspectives of a cornerstone in evolutionary dynamics
Physics of Life Reviews 38, 55 (2021)
Y. C. Ryan, S. E. Ahnert
The Measure of the Archive: The Robustness of Network Analysis in Early Modern Correspondence
Cultural Analytics 7, 57 (2021)
V. Jouffrey, A. S. Leonard, S. E. Ahnert
Gene duplication and subsequent diversification strongly affect phenotypic evolvability and robustness
Royal Society Open Science 8: 201636 (2021)
Y. Ryan, S. Ahnert, R. Ahnert
Networking Archives: Quantitative History and the Contingent Archive
Proceedings of the Workshop on Computational Humanities Research (CHR 2020), http://ceur-ws.org/Vol-2723/ (2020)
S. Cortijo, M. Bhattarai, J. C. W. Locke, S. E. Ahnert
Co-expression Networks From Gene Expression Variability Between Genetically Identical Seedlings Can Reveal Novel Regulatory Relationships
Frontiers in Plant Science 11, 1876 (2020)
R. Ahnert, S. E. Ahnert, C. N. Coleman, S. B. Weingart
The Network Turn: Changing Perspectives in the Humanities
Cambridge University Press (2020) - published Open Access (download)
M. Weiß, S. E. Ahnert
Neutral components show a hierarchical community structure in the genotype-phenotype map of RNA secondary structure
Journal of the Royal Society Interface 17, 20200608 (2020)
M. Weiß, S. E. Ahnert
Using small samples to estimate neutral component size and robustness in the genotype-phenotype map of RNA secondary structure.
Journal of the Royal Society Interface 17, 20190784 (2020)
K. Schiessl, J. L. S. Lilley, T. Lee, I. Tamvakis, W. Kohlen, P. C. Bailey, A. Thomas, J. Luptak, K. Ramakrishnan, M. D. Carpenter, K. S. Mysore, J. Wen, S. Ahnert, V. A. Grieneisen, G. E. D. Oldroyd
NODULE INCEPTION Recruits the Lateral Root Developmental Program for Symbiotic Nodule Organogenesis in Medicago truncatula
Current Biology 29, 1 (2019)
J. Zhang, G. Eswaran, J. Alonso-Serra, M. Kucukoglu, J. Xiang, W. Yang, A. Elo, K. Nieminen, T. Damén, J.-G. Joung, J.-Y. Yun, J.-H. Lee, L. Ragni, P. Barbier de Reuille, S. E. Ahnert, J.-Y. Lee, A. P. Mähönen, Y. Helariutta
Transcriptional regulatory framework for vascular cambium development in Arabidopsis roots
Nature Plants 5, 1033 (2019)
E. K. Towlson, P. E. Vértes, U. Müller-Sedgwick, S. E. Ahnert
Brain Networks Reveal the Effects of Antipsychotic Drugs on Schizophrenia Patients and Controls
Frontiers in Psychiatry 10, 611 (2019)
A. S. Leonard, S. E. Ahnert
Evolution of interface binding strengths in simplified model of protein quaternary structure
PLOS Computational Biology 15(6): e1006886 (2019)
S. Cortijo, Z. Aydin, S. E. Ahnert, J. C. W. Locke
Widespread inter-individual gene expression variability in Arabidopsis thaliana
Molecular Systems Biology 15: e8591 (2019)
R. Ahnert, S. E. Ahnert
Metadata, Surveillance and the Tudor State
History Workshop Journal 87, 27 (2019)
V. Sekara, P. Deville, S. E. Ahnert, A.-L. Barabási, R. Sinatra, S. Lehmann
The chaperone effect in scientific publishing
Proceedings of the National Academy of Sciences 115, 12603 (2018)
S. Tesoro, S. E. Ahnert
Non-deterministic genotype-phenotype maps of biological self-assembly
Europhysics Letters 123, 38002 (2018)
W. P. Grant, S. E. Ahnert
Modular decomposition of protein structure using community detection
Journal of Complex Networks, cny014 (2018)
S. Tesoro, S. E. Ahnert, A. S. Leonard
Determinism and boundedness of self-assembling structures
Physical Review E 98, 022113 (2018)
M. Ikeuchi, M. Shibata, B. Rymen, A. Iwase, A.-M. Bågman, L. Watt, D. Coleman, D. S. Favero, T. Takahashi, S. E. Ahnert, S. M. Brady, K. Sugimoto
A Gene Regulatory Network for Cellular Reprogramming in Plant Regeneration
Plant and Cell Physiology, pcy013 (2018)
M. Weiß, S. E. Ahnert
Phenotypes can be robust and evolvable if mutations have non-local effects on sequence constraints
Journal of the Royal Society Interface 15, 20170618 (2018)
S. E. Ahnert, W. P. Grant, C. J. Pickard
Revealing and exploiting hierarchical material structure through complex atomic networks
npj Computational Materials 3, 35 (2017)
G. F. Chami, S. E. Ahnert, N. B. Kabatereine, E. M. Tukahebwa
Social network fragmentation and community health
Proceedings of the National Academy of Sciences 114, E7425 (2017)
S. E. Ahnert
Structural properties of genotype-phenotype maps
Journal of the Royal Society Interface 14, 20170275 (2017)
O. G. Mouritsen, R. Edwards-Stuart, Y.-Y. Ahn, S. E. Ahnert
Data-driven Methods for the Study of Food Perception, Preparation, Consumption, and Culture
Frontiers in ICT 4, 15 (2017)
A. S. Fokas, D. J. Cole, S. E. Ahnert, A. W. Chin
Residue Geometry Networks: A Rigidity-Based Approach to the Amino Acid Network and Evolutionary Rate Analysis
Scientific Reports 6, 33213 (2016)
S. Tesoro, K. Göpfrich, T. Kartanas, U. F. Keyser, S. E. Ahnert
Nondeterministic self-assembly with asymmetric interactions
Physical Review E 94, 022404 (2016)
S. E. Ahnert, T. M. A. Fink
Form and function in gene regulatory networks: the structure of network motifs determines fundamental properties of their dynamical state space
Journal of the Royal Society Interface 13, 20160179 (2016)
S. Tesoro, S. E. Ahnert
Nondeterministic self-assembly of two tile types on a lattice
Physical Review E 93, 042412 (2016) - selected as Editors' Suggestion
S. F. Greenbury, S. Schaper, S. E. Ahnert, A. A. Louis
Genetic correlations greatly increase mutational robustness and can both reduce and enhance evolvability
PLOS Computational Biology 12(3): e1004773 (2016)
S. E. Ahnert, J. A. Marsh, H. Hernandez, C. V. Robinson, S. A. Teichmann
Principles of assembly reveal a periodic table of protein complexes
Science 350, 1331 (2015)
S. F. Greenbury, S. E. Ahnert
The organization of biological sequences into constrained and unconstrained parts determines fundamental properties of genotype-phenotype maps
Journal of The Royal Society Interface 12, 20150724 (2015)
R. Ahnert, S. E. Ahnert
Protestant letter networks in the reign of Mary I: A quantitative approach
English Literary History 82, 1 (2015)
J. Marsh, H. Rees, S. E. Ahnert, S. A. Teichmann
Structural and evolutionary versatility in protein complexes with uneven stoichiometry
Nature Communications 6, 6394 (2015)
M. Taylor-Teeples, L. Lin, M. de Lucas M, G. Turco, T. W. Toal, A. Gaudinier, N. F. Young, G. M. Trabucco, M. T. Veling, R. Lamothe, P. P. Handakumbura, G. Xiong, C. Wang, J. Corwin, N. Tsoukalas, L. Zhang, D. Ware, M. Pauly, D. J. Kliebenstein, K. Dehesh, I. Tagkopoulos, G. Breton, J. Pruneda-Paz, S. E. Ahnert, S. A. Kay, S. P. Hazen, S. M. Brady
An Arabidopsis Gene Regulatory Network for Xylem Specification and Secondary Wall Biosynthesis
Nature 517, 571 (2015)
S. Shin, S. E. Ahnert, J. Park
Ranking Competitors Using Degree-Neutralized Random Walks
PLOS ONE 9(12): e113685 (2014)
G. F. Chami, S. E. Ahnert, M. J. Voors, A. A. Kontoleon
Social Network Analysis Predicts Health Behaviours and Self-Reported Health in African Villages
PLOS ONE 9(7): e103500 (2014)
R. Ahnert, S. E. Ahnert
A community under attack: Protestant letter networks in the reign of Mary I
Leonardo 47, 275 (2014)
S. E. Ahnert
Generalised power graph compression reveals dominant relationship patterns in complex networks
Scientific Reports 4, 4385 (2014)
S. F. Greenbury, I. G. Johnston, A. A. Louis, S. E. Ahnert
A tractable genotype-phenotype map modelling the self-assembly of protein quaternary structure
Journal of The Royal Society Interface 11, 20140249 (2014)
S. E. Ahnert
Power graph compression reveals dominant relationships in genetic transcription networks
Molecular BioSystems 9, 2681 (2013)
D. Garlaschelli, S. E. Ahnert, T. Fink, G. Caldarelli
Low-Temperature Behaviour of Social and Economic Networks
Entropy 15, 3148 (2013)
Y.-Y. Ahn, S. E. Ahnert
The Flavor Network
Leonardo 46, 272 (2013)
J. A. Marsh, H. Hernandez, Z. Hall, S. E. Ahnert, T. Perica, C. V. Robinson, S. A. Teichmann
Protein complexes are under evolutionary selection to assemble via ordered pathways
Cell 153, 461 (2013)
E. K. Towlson, P. Vertes, S. E. Ahnert, W. Schafer, E. Bullmore
The rich club of the C. elegans neuronal connectome
Journal of Neuroscience 33, 6380 (2013)
S. E. Ahnert
Network analysis and data mining in food science: the emergence of computational gastronomy
Flavour 2:4 (2013)
T. Perica, J. A. Marsh, F. L. Sousa, E. Natan, L. J. Colwell, S. E. Ahnert, S. A. Teichmann
The emergence of protein complexes: quaternary structure, dynamics and allostery
Biochem. Soc. Trans. 40, 475 (2012)
Y. Y. Ahn, S. E. Ahnert, J. P. Bagrow, A.-L. Barabási
Flavor network and the principles of food pairing
Scientific Reports 1:196 (2011)
I. G. Johnston, S. E. Ahnert, J. P. K. Doye, A. A. Louis
Evolutionary dynamics in a simple model of self-assembly
Physical Review E 83, 066105 (2011)
S. M. Brady, L. Zhang, M. Megraw, N. J. Martinez, E. Jiang, C. S. Yi, W. Liu, A. Zeng, M. Taylor-Teeples, D. Kim, S. E. Ahnert, U. Ohler, D. Ware, A. J. M. Walhout, P. N. Benfey
A stele-enriched gene regulatory network in the Arabidopsis root
Molecular Systems Biology, 7:459 (2011)
M. A. Moreno-Risueno, J. M. Van Norman, A. Moreno, J. Zhang, S. E. Ahnert, P. N. Benfey
Oscillating Gene Expression Determines Competence for Periodic Arabidopsis Root Branching
Science 329, 1306 (2010)
S. E. Ahnert, I. G. Johnston, T. M. A. Fink, J. P. K. Doye, A. A. Louis
Self-assembly, modularity and physical complexity
Physical Review E 82, 026117 (2010)
D. A. Orlando, S. M. Brady, T. M. A. Fink, P. N. Benfey, S. E. Ahnert
Detecting separate time scales in genetic expression data
BMC Genomics 11:381 (2010)
S. E. Ahnert, B. A. N. Travencolo, L. da Costa Fontoura
Connectivity and dynamics of neuronal networks as defined by the shape of individual neurons
New Journal of Physics 11, 103053 (2009)
J. B. Coe, S. E. Ahnert, T. M. A. Fink
When are cellular automata random?
Europhysics Letters 84, 50005 (2008)
S. E. Ahnert, S. A. Teichmann
Networks for all
Genome Biology 9, 324 (2008)
S. E. Ahnert, T. M. A. Fink
Clustering signatures classify directed networks
Physical Review E 78, 036112 (2008)
T. M. A. Fink, J. B. Coe, S. E. Ahnert
Single-elimination competition
Europhysics Letters 83, 60010 (2008)
M.-L. Dequeant, S. E. Ahnert, H. Edelsbrunner, T. M. A. Fink, Y. Mileyko, J. Morton, A. R. Mushegian, L. Pachter, M. Rowicka, A. Shiu, B. Sturmfels, O. Pourquie
Comparison of Pattern Detection Methods in Microarray Time Series of the Segmentation Clock
PLOS ONE 3(8): e2856 (2008)
S. E. Ahnert, D. Garlaschelli, T. M. A. Fink, G. Caldarelli
Applying weighted network measures to microarray distance matrices
Journal of Physics A 41, 224011 (2008)
S. E. Ahnert, T. M. A. Fink, A. Zinovyev
How much non-coding DNA do eukaryotes require?
Journal of Theoretical Biology 252, 587 (2008)
S. E. Ahnert, D. Garlaschelli, T. M. A. Fink, G. Caldarelli
An ensemble approach to the analysis of weighted networks
Physical Review E 76, 016101 (2007)
S. E. Ahnert, K. Willbrand, F. C. S. Brown, T. M. A. Fink
Unbiased pattern detection in microarray data series
Bioinformatics 22, 1471 (2006)
S. E. Ahnert, M. C. Payne
All possible bipartite positive-operator-value measurements of two-photon polarization states
Physical Review A 73, 022333 (2006)
S. E. Ahnert, M. C. Payne
General implementation of all possible positive-operator-value measures of single photon polarization states
Physical Review A 71, 012330 (2005)
S. E. Ahnert, M. C. Payne
Linear optics implementation of weak values in Hardy's paradox
Physical Review A 70, 042102 (2004)
S. E. Ahnert, M. C. Payne
Weak measurement of the arrival times of single photons and pairs of entangled photons
Physical Review A 69, 042103 (2004)
S. E. Ahnert, M. C. Payne
Nonorthogonal projective positive-operator-value measurement of photon polarization states with unit probability of success
Physical Review A 69, 012312 (2004)






