Submitted by Ellie Hall on Mon, 10/01/2022 - 08:33
Researchers from our Laser Analytics and Molecular Neuroscience groups used advanced super-resolution microscopy methods to visualise the movements of four proteins that are key to the replication cycle of the SARS-CoV-2 virus within host cells. The work, published in Science Advances, furthers our understanding of how the virus behind COVID-19 operates.
The SARS-CoV-2 RNA virus is the causative agent of COVID-19, a disease that has spawned a global pandemic with more than 239 million cases diagnosed worldwide. While new diagnosis, prevention, and treatment options for COVID-19 continue to emerge at a rapid pace, our understanding of the fundamental biology of the SARS-CoV-2 virus itself has advanced more slowly.
Unravelling the mechanisms of transmission and replication of this virus is crucial for the informed development of drugs and vaccines, and to understand the long-term effects of the disease, allowing researchers to better develop countermeasures against evolving variants of concern.
“We have been using fluorescence microscopy to look at the replication cycle of different viruses, such as herpes and influenza,” says Dr Katharina Scherer, a postdoctoral researcher in the Laser Analytics group at CEB. “We knew a lot about how to image virus replication in cells with the advanced, modern light microscopes we have in the department, and also how to develop new methods to image specific processes within the replication cycle.
“So, when the coronavirus pandemic started, we were looking for a way to contribute.”
When the SARS-CoV-2 virus enters your body, it binds to cells in your respiratory tract and releases a strand of material called RNA that tells the cells to produce a number of proteins that can be combined together to make new virus particles. These four proteins are known as structural proteins – they support the overall structure of a new virus particle and are therefore often good targets for drugs looking to prevent or minimise infection.
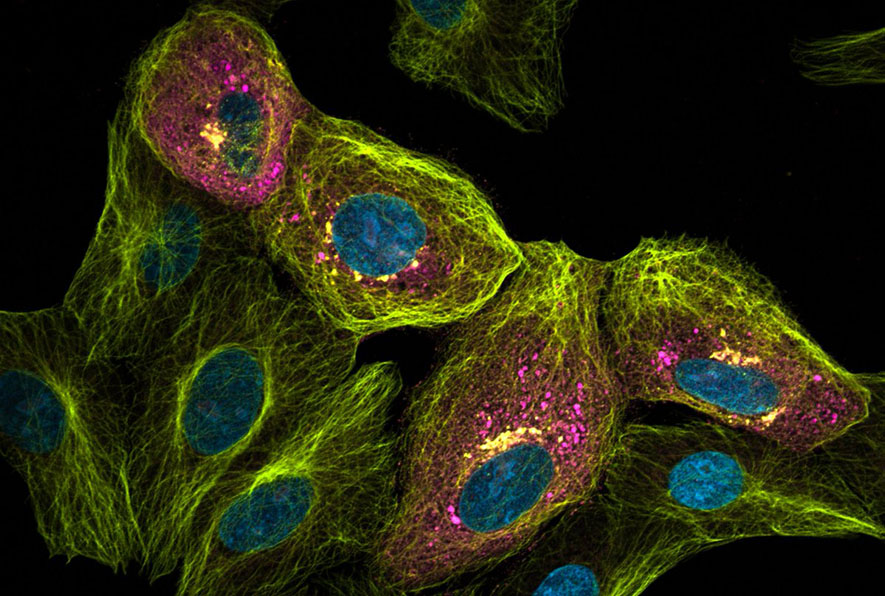
Image: Sars Cov-2 infected cell visualised with optical super resolution imaging. Virus load is shown in magenta, the microtubules of the cells are shown in yellow and the nucleus in cyan.
“We wanted to know more about the SARS-CoV-2 life cycle; to study the structural proteins that are created in the host cell at different times and in different places and how they come together to form the new virus particles that are then released from the cell into the body,” says Scherer.
“This is useful to know in detail because if you want to develop a therapeutic for the virus, you want to know where you can target – what are the sensitive steps, where are the bottlenecks that you can potentially attack with a drug.”
Using samples of cells that had been infected with SARS-CoV-2 by Professor Jonathan Heeney’s research group at the Department of Veterinary Medicine in their Containment Level 3 (CL3) laboratory, Scherer, Mascheroni and colleagues applied sophisticated microscopy methods to visualise the movements of these four key structural proteins throughout the host cell at various stages of infection.
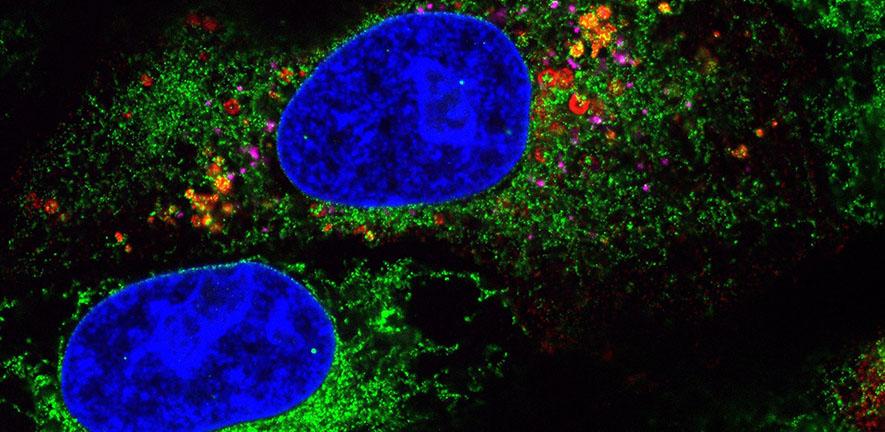
Their method of combining expansion microscopy with light sheet microscopy enabled them to generate much higher resolution images of a single cell, allowing them to look at several different proteins and features at the same time – something that is not achievable with other super-resolution microscopy techniques. This allowed them to see exactly where the different structural proteins are in relation to each other and other components within the cell, providing much more detail on what is happening at the various stages of infection and virus replication.
They found a number of differences in the expression of the structural proteins and how they move through the cell to facilitate virus replication. Three of the proteins are within the membranes and outer layers of the virus and the fourth wraps up the viral genome – all of the essential genetic information that the virus needs to be able to replicate itself.
This fourth protein, the so-called nucleocapsid protein is produced very early on, before the other three proteins, and in a different location within the cell. The nucleocapsid protein accumulates very close to material in the cell that it can use to protect the virus’s genetic information and this likely enables it to wrap up this viral genome very quickly, before it can be detected by the cell’s immune system.
The nucleocapsid protein together with the genome is then transported to where the other three structural proteins are, so that together, they can form a new virus particle that can be released from the cell.
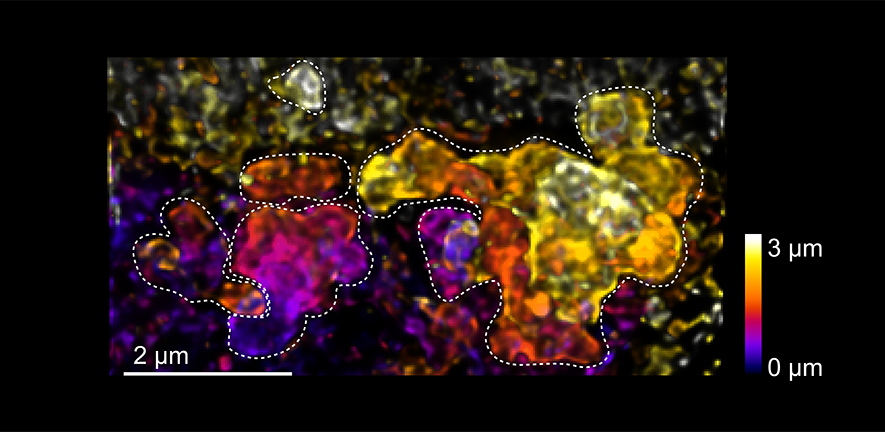
Image: microscopy image showing how the genome-packaging protein of SARS-CoV-2 accumulates at virus-made membrane compartments (dashed lines). Inside the virus replicates its genome.
The virus uses many of the host cell’s own structures to facilitate its replication – such as carrier vesicles called lysosomes to transport newly replicated viruses out of the cell. Using these microscopy techniques allowed Scherer, Mascheroni and colleagues to not only visualise changes in the viruses’ structural proteins, but also see changes in the host cell’s own structures in relation to these.
“All of this helps us understand the conditions under which replication is easy and what conditions may make it more difficult for the virus to replicate,” says Scherer.
“It gives you information to see what may be good treatment strategies and what are not. For example, we see that the nucleocapsid protein binds the virus genome very early and immediately before it can be discovered by the immune system. This is useful to know because it tells you that attacking the genome of the virus is going to be difficult - it is protected very quickly in these vesicles and is not really exposed to the cell.
“So maybe you can think of strategies to attack the nucleocapsid protein instead of the genetic material itself.”
The group are now applying the same techniques to the different variants of SARS-CoV-2 to understand also why different variants behave differently and why they may be more or less pathogenic than others.
They are also planning to apply these techniques to visualise the effects of potential drug compounds that could target RNA binding proteins involved in SARS-CoV-2 replication and hope it will be a useful validation method for any results from these studies.
“I am proud of what we have achieved in this collaboration,” said Professor Clemens Kaminski, head of the Laser Analytics research group. “By bringing in the expertise from the Molecular Neuroscience and Heeney groups in CEB, and the Vet School, we were in an ideal position to gain new insights on the assembly process of SARS-CoV-2 in the cell.
“The images Luca and Katharina have produced are beautiful to look at and provide fascinating new insights into workings of this deadly virus. An improved understanding of its replication cycle may lead to the development of completely new therapeutic strategies."
Read the full paper: Scherer K. et al: ‘SARS-CoV-2 nucleocapsid protein adheres to replication organelles before viral assembly at the Golgi/ERGIC and lysosome-mediated egress’, Science Advances, January 2022 DOI 10.1126/sciadv.abl4895