Examples of current projects include:
Monitoring of Viral Activity using Plasma Membrane - Conducting Polymer Chip Assembly
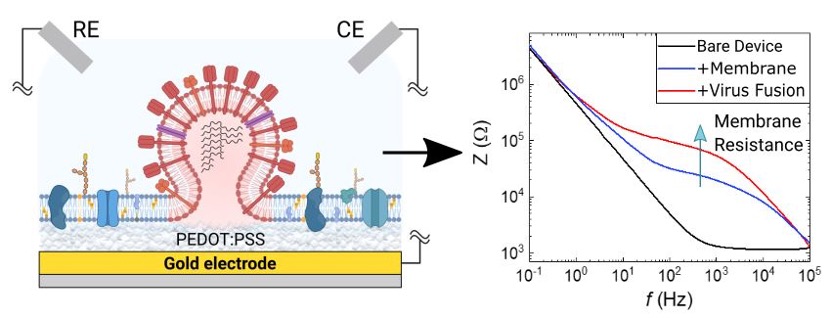
The majority of anti-viral drugs prevent cellular infection by acting on proteins found on host cell membranes. In this project we couple native plasma membranes with organic electronic chips to create biosensors that can distinguish between the different stages of viral entry. Our platform uses a label-free electronic readout, combined with conventional optical readouts (fluorescence) to sense the ability of viruses to bind and fuse with host cell membranes, thereby sensing viral entry. Once a virus fuses with our membrane-chip assembly, its membrane merges with the plasma membrane, further inhibiting ion injection to the conducting polymer film. Our organic electronic chips then transduce this additional ionic barrier to electronic output via conventional electrochemical methods such as electrochemical impedance spectroscopy (EIS). This platform has been validated for different viruses including influenza and SARS-CoV-2, while our end goal is to develop it into a drug-screening tool to assist and re-direct the drug discovery process. This project is funded by the US military agency (DARPA) and is the product of collaboration between the Daniel group at Cornell University and the Salleo group at Stanford University.
Innovative technology solutions to explore effects of the microbiome on intestine and brain pathophysiology
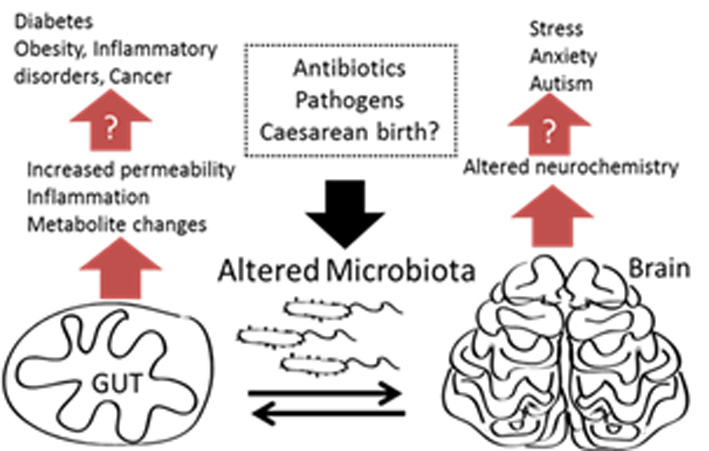 In this project we are generating in vitro models of the gut and brain with integrated monitoring based on our organic electronics device toolbox.[2–4] The completed model will be used to monitor effects of the microbiome on the gut brain axis. We are building on former expertise in generating models of the gut for pathogen detection,[5,6] and also expertise in generating a 3D models of different tissues[7] including the blood brain barrier for toxicology experiments. Also interfacing with this work is our ability to generate multi-electrode arrays from conducting polymers for monitoring neuronal activity from 2D and 3D cultures. This is an ERC funded project which started on 1/10/2017.
In this project we are generating in vitro models of the gut and brain with integrated monitoring based on our organic electronics device toolbox.[2–4] The completed model will be used to monitor effects of the microbiome on the gut brain axis. We are building on former expertise in generating models of the gut for pathogen detection,[5,6] and also expertise in generating a 3D models of different tissues[7] including the blood brain barrier for toxicology experiments. Also interfacing with this work is our ability to generate multi-electrode arrays from conducting polymers for monitoring neuronal activity from 2D and 3D cultures. This is an ERC funded project which started on 1/10/2017.
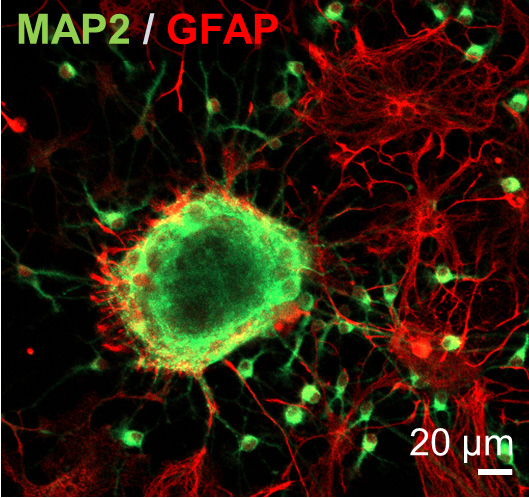
Confocal image of 3D neurospheres stained for astrocytes (red) and neurons (green). From: Pas et al, "Enhancing electrophysiology recordings with neurospheres via patterned PEDOT:PSS microelectrode arrays”. Advanced Biosystems (2017) doi: 10.1002/adbi.201700164. Copyright Wiley-VCH Verlag GmbH & Co. KGaA. Reproduced with permission.
Tissue engineering using composite conducting polymer/bio- polymer scaffolds
We have demonstrated the use of electroactive conducting polymer scaffolds for hosting monitoring cells.[8] These scaffolds are easily blended with biopolymers such as collagen which can be used as a tool to modulate biochemical and mechanical properties of the scaffolds. Thanks to their electrical properties, the scaffolds can be used to stimulate cells electrically which has been shown to alter/enhance differentiation and proliferation of cells under certain conditions.
Cover image illustrating conducting polymer scaffolds used for co-culture of mammalian cells. The cover was designed by Heno Hwang, scientific illustrator, King Abdullah University of Science and Technology. From: Inal et al, "Conducting Polymer Scaffolds for Monitoring 3D Cell Culture”. Advanced Biosystems 1 (6); 1700052 (2017) doi: 10.1002/adbi.201700052. Copyright Wiley-VCH Verlag GmbH & Co. KGaA. Reproduced with permission.

Integration of lipid monolayers and bilayers with organic electronic devices for drug screening
Lipid bilayers or monolayers may be used as a surrogate system to understand interactions happening at the outside of bacterial or mammalian cells. There has been a longstanding interest in the bioelectronics community to efficiently interface lipids with optical and electronic transducers. We have shown that lipid bilayers can be integrated with organic electronic devices for monitoring ion channel function.[9] This is combined with our fundamental interest in understanding the nature of the interface of our devices with biological materials that exist on the outer layer of a cell such as proteins, sugars and lipids.[10] As well as bilayers we can also generate monolayers on our devices and study interactions between compounds and lipids which strongly affect electrical properties of our devices. This is a collaborative project with Profs Daniel and Alabi at Cornell University and Prof. Salleo at Stanford University. We are using innovative device design to generate an electrical monitoring system which gives highly quantitative data on disruption of bacterial lipid membranes by antibacterial compounds.
Metabolite sensing from live cells
A long running project in the group has been highly sensitive and specific metabolite sensing in complex biological samples, including from live cells. Biofunctionalisation of our conducting polymer devices[11] is a fundamental of this research activity and it feeds into our other activities as a means to render devices more biocompatible, integrate additional functionalities such as metabolite sensing or even change the physicochemical properties of our materials to enhance biological phenomena. We have demonstrated metabolite sensing from live cells under stress[12], from saliva samples of volunteers,[13] and most recently from tumour cell cultures to predict metastatic potential[14]. The goal here is to integrate the metabolite sensing into our organ on chip projects to generate multi-parameter analysis of cell function in a dynamic and continuous manner.
References
[1] J. Rivnay, R. M. Owens, G. G. Malliaras, Chem. Mater. 2014, 26, 679.
[2] V. F. Curto, B. Marchiori, A. Hama, A.-M. Pappa, M. P. Ferro, M. Braendlein, J. Rivnay, M. Fiocchi, G. G. Malliaras, M. Ramuz, R. M. Owens, 2017, 3, 17028.
[3] J. Rivnay, P. Leleux, A. Hama, M. Ramuz, M. Huerta, G. G. Malliaras, R. M. Owens, Sci. Rep. 2015, 5, 11613.
[4] M. Huerta, J. Rivnay, M. Ramuz, A. Hama, R. M. Owens, Appl. Vitro Toxicol. 2016, 2, 17.
[5] S. A. Tria, L. H. Jimison, A. Hama, M. Bongo, R. M. Owens, Biochim Biophys Acta 2013, 1830, 4381.
[6] S. A. Tria, M. Ramuz, M. Huerta, P. Leleux, J. Rivnay, L. H. Jimison, A. Hama, G. G. Malliaras, R. M. Owens, Adv Heal. Mater 2014, 3, 1053.
[7] M. Huerta, J. Rivnay, M. Ramuz, A. Hama, R. M. Owens, APL Mater. 2015, 3, 030701.
[8] S. Inal, A. Hama, M. Ferro, C. Pitsalidis, J. Oziat, D. Iandolo, A.-M. Pappa, M. Hadida, M. Huerta, D. Marchat, P. Mailley, R. M. Owens, Adv. Biosyst. 2017, 1700052.
[9] Y. Zhang, S. Inal, C.-Y. Hsia, M. Ferro, M. Ferro, S. Daniel, R. M. Owens, Adv. Funct. Mater. 2016, 26, 7304.
[10] A.-M. Pappa, S. Inal, K. Roy, Y. Zhang, C. Pitsalidis, A. Hama, J. Pas, G. G. Malliaras, R. M. Owens, ACS Appl. Mater. Interfaces 2017, 9, 10427.
[11] X. Strakosas, M. Sessolo, A. Hama, J. Rivnay, E. Stavrinidou, G. G. Malliaras, R. M. Owens, J. Mater. Chem. B 2014, 2, 2537.
[12] X. Strakosas, M. Huerta, M. J. Donahue, A. Hama, A.-M. Pappa, M. Ferro, M. Ramuz, J. Rivnay, R. M. Owens, J. Appl. Polym. Sci. 2017, 134, DOI 10.1002/app.44483.
[13] A.-M. Pappa, V. F. Curto, M. Braendlein, X. Strakosas, M. J. Donahue, M. Fiocchi, G. G. Malliaras, R. M. Owens, Adv. Healthc. Mater. 2016, 5, 2295.
[14] M. Braendlein, A.-M. Pappa, M. Ferro, A. Lopresti, C. Acquaviva, E. Mamessier, G. G. Malliaras, R. M. Owens, Adv. Mater. 2017, 1605744.